
Bioinformatics | Big data from tiny ocular fluids | Tear Film | Aqueous Humor | Vitreous Fluid | Biomarkers| Ocular Surface

Ashok Sharma, PhD
Associate Professor
Genomic Medicine Graduate Program Director
Center for Biotechnology & Genomic Medicine
Department of Ophthalmology
Medical College of Georgia at Augusta University
706-721-6335
Jump to: Education Research Interests Media Articles & Publications Graduate Students & Staff
With a background in computer science and a PhD in Genomic Medicine, I have experience in both the computational and biological aspects of biomedical research. My research focus is on the development of biologically meaningful algorithms for comprehensive analysis and the visualization of high-throughput data by incorporating biological knowledge and rigorous computational methods. In the last decade, biomedical research has been revolutionized by the advent of high-throughput technologies which can generate large amounts of data. Unlocking the meaningful biological information contained inside high-throughput data is a big challenge for bench biologists. In order to facilitate the analysis of such complex data, it is crucial to develop statistical tools as well as efficient algorithms and methods to extract useful information. My primary area of research is bioinformatics, where I apply computational, mathematical, and statistical methods to solve complex biological problems. Long term goals of my lab are to solve the challenges in the analysis of data from many high-throughput technologies, especially mass spectrometry-based proteomics, and to implement these solutions in usable software tools.
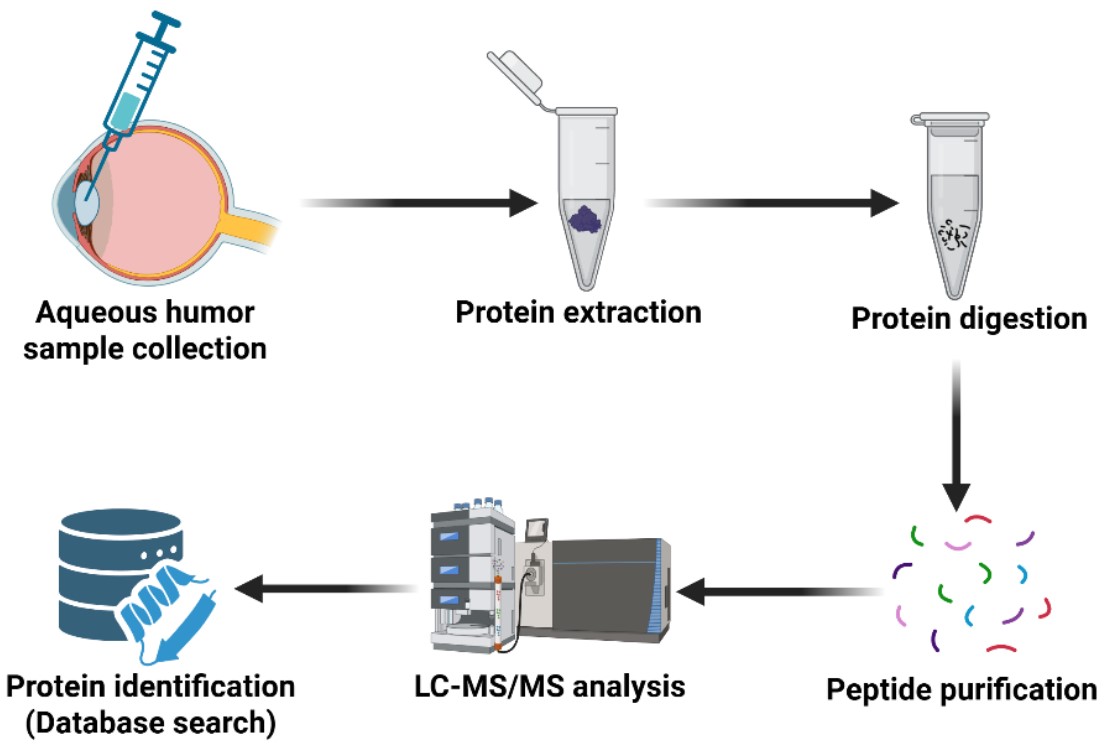
Aqueous humor (AH) is the fluid in the anterior and posterior chambers of the eye that contains proteins regulating many ocular health functions, including nutrient and oxygen supply, the removal of metabolic waste, ocular immunity, and ocular shape and refraction. The dynamics of AH and the fine balance between production and drainage is essential in maintaining the physiological intraocular pressure (IOP). Therefore, identifying the protein contents of AH is vital in understanding their physiological and pathological roles in the eye.
By utilizing a large sample set, state of the art technology, and revolutionary data analysis methods, we identified the constitutive proteome of human aqueous humor, which may be useful as a reference for future studies. Discovery of AH proteomic alterations associated with glaucomatous optic neuropathy and glaucoma risk factors will help the research community at large in understanding physiological and pathological proteomics signatures in the AH.
Website: https://ahp.augusta.edu/
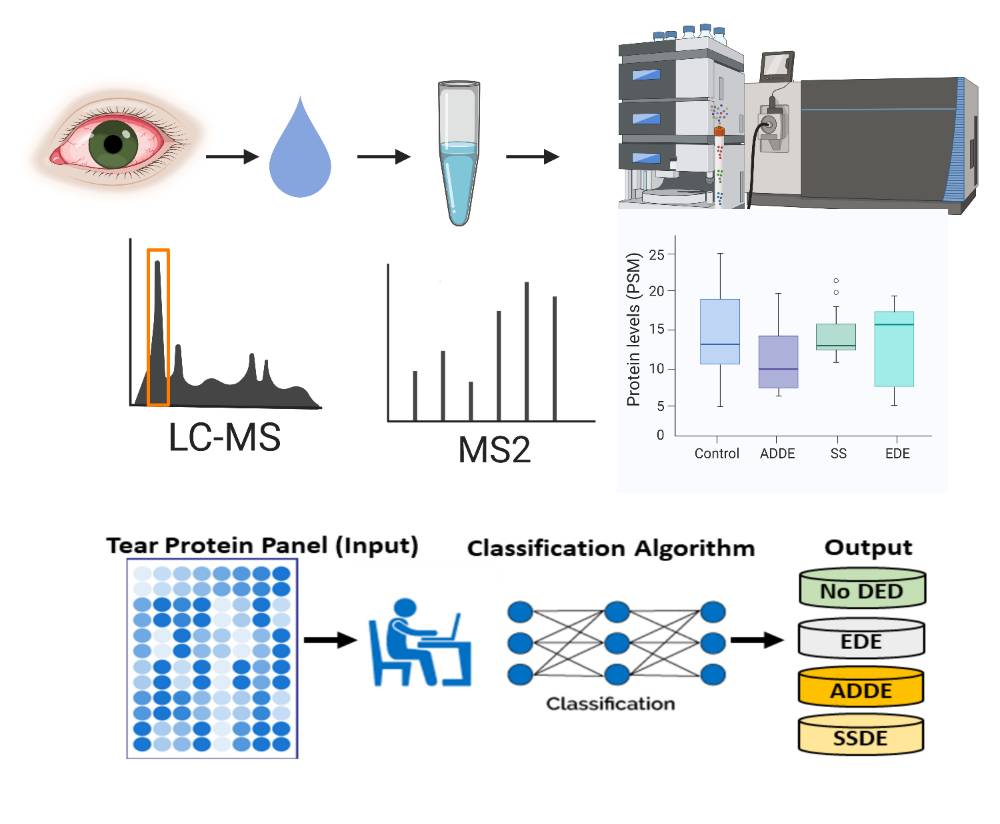
The tear film is a thin (2-6 µm) multi-layer fluid that coats the corneal and conjunctival epithelia. The proximity of the tear film to the ocular milieu, the non-invasive nature of its collection and its high protein concentration make it an attractive source for proteomic biomarker studies. We have developed a new workflow for proteomic analysis of tear fluid samples obtained with Schirmer strips that can identify over 3000 proteins in tear samples from human subjects.
We plan to collect a large number of tear samples using a standardized method and then, develop a reference tear proteomic database to prioritize, integrate, and analyze potential biomarkers. This will provide a readily accessible and permanent resource, fostering collaboration between laboratories to expand and improve the database. It is also likely that this database will help the vision research community to understand the physiological and pathological proteomic signatures in tear fluid.
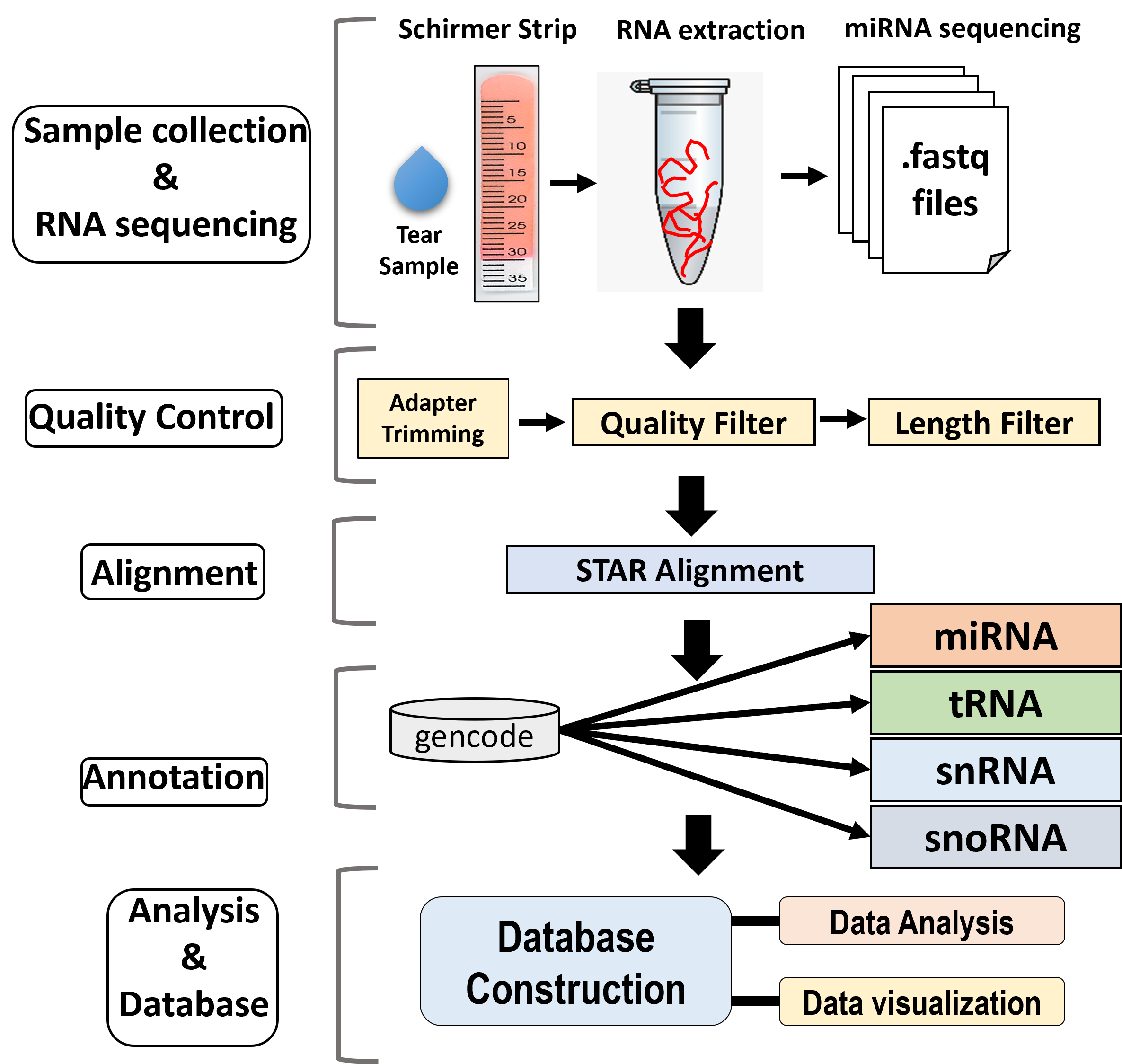
Small non-coding RNAs (sncRNAs) are diverse RNA molecules with potential applications as biomarkers and therapeutic targets in various diseases due to their role in gene expression regulation. Subtypes, including microRNA (miRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), and transfer RNA (tRNA), have distinct functions and lengths. Recent research highlights the presence of extracellular RNA in human tears, showing altered sncRNA levels in ocular disorders (e.g., diabetic retinopathy, glaucoma, and DED) and non-ocular diseases (e.g., Alzheimer’s disease, breast cancer). Despite being in its early stages, this project aims to bridge the knowledge gap by leveraging advanced technologies and software to analyze tear sncRNAs. The findings will lay the groundwork for future studies using tear biomarkers to enhance diagnostic methods.
I work with an interdisciplinary team to carry out bioinformatics analysis and to pursue research projects in collaboration with other biologists. Currently, I am developing analytical methods for the large-scale measurements that are part of several Cancer and Diabetes projects. In our center (CBGM: Center for Biotechnology and Genomic Medicine), I am providing bioinformatics support for several long-term studies including The Environmental Determinants of Diabetes in the Young (TEDDY), the Phenome and Genome of Diabetes Autoimmunity (PAGODA), the Diabetic Complications Consortium (DiaComp), the Mouse Metabolic Phenotype Consortium (MMPC) and Biomarkers and Therapeutics in Cancer (BAT Cancer).
Spectrometry Analyses
Vision Research Projects

MCG scientists establish protein database to advance vision research
Scientists at the Medical College of Georgia at Augusta University have established a database that will allow researchers to better examine the underlying causes of some of the most common conditions that cause vision loss.

Noninvasive technique collects sufficient tear fluid to look for biomarkers of health and disease
The protective outer layer of our eyes, called the tear film, contains thousands of proteins, which provide clues about wellness and disease, and scientists have fine-tuned what they say is a non-invasive and efficient way to look at those clues.

Genetic discovery may help better identify children at risk for type 1 diabetes
Six novel chromosomal regions identified by scientists leading a large, prospective study of children at risk for type 1 diabetes will enable the discovery of more genes that cause the disease and more targets for treating or even preventing it.
New technology enables identification of biomarkers for a wide range of diseases
Genetic discovery may help better identify children at risk for type 1 diabetes
Powerful anti-inflammatory molecule may block vision loss in diabetic retinopathy
Selected publications:
Williams E, Altman J, Ahmed S, Jones G, Khadanga A, Alevy D, Bollinger K, Estes A, Safille S, Lee TJ, Sharma S, Sharma A. Unveiling the levels and significance of different serpin family proteins in aqueous humor dynamics. BMC Ophthalmol. 2025 May 19;25(1):297. doi: 10.1186/s12886-025-04119-3. PubMed PMID: 40389884; PubMed Central PMCID: PMC12090511.
Lee TJ, Goyal A, Jones G, Glass J, Doshi V, Bollinger K, Ulrich L, Ahmed S, Kodeboyina SK, Estes A, Töteberg-Harms M, Zhi W, Sharma S, Sharma A. AHP DB: a reference database of proteins in the human aqueous humor. Database (Oxford). 2024 Jan 29;2024. doi: 10.1093/database/baae001. PubMed PMID: 38284936; PubMed Central PMCID: PMC10878049.
Jones G, Altman J, Ahmed S, Lee TJ, Zhi W, Sharma S, Sharma A. Unraveling the Intraday Variations in the Tear Fluid Proteome. Invest Ophthalmol Vis Sci. 2024 Mar 5;65(3):2. doi: 10.1167/iovs.65.3.2. PubMed PMID: 38441890; PubMed Central PMCID: PMC10916888.
Beisel A, Jones G, Glass J, Lee TJ, Töteberg-Harms M, Estes A, Ulrich L, Bollinger K, Sharma S, Sharma A. Comparative analysis of human tear fluid and aqueous humor proteomes. Ocul Surf. 2024 Mar 30;. doi: 10.1016/j.jtos.2024.03.011. [Epub ahead of print] PubMed PMID: 38561100.
Ahmad Z, Singh S, Lee TJ, Sharma A, Lydic TA, Giri S, Kumar A. Untargeted and temporal analysis of retinal lipidome in bacterial endophthalmitis. Prostaglandins Other Lipid Mediat. 2024 Apr;171:106806. doi: 10.1016/j.prostaglandins.2023.106806. Epub 2024 Jan 5. Review. PubMed PMID: 38185280; PubMed Central PMCID: PMC10939753.
Lee H, Lee TJ, Galloway CA, Zhi W, Xiao W, de Mesy Bentley KL, Sharma A, Teng Y, Sesaki H, Yoon Y. The mitochondrial fusion protein OPA1 is dispensable in the liver and its absence induces mitohormesis to protect liver from drug-induced injury. Nat Commun. 2023 Oct 23;14(1):6721. doi: 10.1038/s41467-023-42564-0. PubMed PMID: 37872238; PubMed Central PMCID: PMC10593833.
Vashishtha A, Maina SW, Altman J, Jones G, Lee TJ, Bollinger KE, Ulrich L, Töteberg-Harms M, Estes AJ, Zhi W, Sharma S, Sharma A. Complement System Proteins in the Human Aqueous Humor and Their Association with Primary Open-Angle Glaucoma. J Pers Med. 2023 Sep 19;13(9). doi: 10.3390/jpm13091400. PubMed PMID: 37763167; PubMed Central PMCID: PMC10532607.
Altman J, Jones G, Ahmed S, Sharma S, Sharma A. Tear Film MicroRNAs as Potential Biomarkers: A Review. Int J Mol Sci. 2023 Feb 12;24(4). doi: 10.3390/ijms24043694. Review. PubMed PMID: 36835108; PubMed Central PMCID: PMC9962948.
Jones G, Lee TJ, Glass J, Rountree G, Ulrich L, Estes A, Sezer M, Zhi W, Sharma S, Sharma A. Comparison of Different Mass Spectrometry Workflows for the Proteomic Analysis of Tear Fluid. Int J Mol Sci. 2022 Feb 19;23(4). doi: 10.3390/ijms23042307. PubMed PMID: 35216421; PubMed Central PMCID: PMC8875482.

Saleh Ahmed
706-721-3515


Garrett N. Jones
706-836-9752

Jeremy Altman
706-721-3515


Sharon W. Maina
706-721-3515





