- Augusta University
- Research
- Core Laboratories
- Proteomics Core
Proteomics Core
The Proteomics Core at Augusta University is a resource facility for the characterization and expression level quantitation of proteins (isolated or in an extract) and small molecules/metabolites by mass spectrometry (MS) and/or chromatography.
Our Mission
Our Goal
While the Proteomics Core is a fee-for-service facility, a major goal is to become a research environment for multidisciplinary research and education that utilizes mass spectrometry and other proteomics technologies to resolve complex biological and medical problems.
The Proteomics Core is operated by the Center for Biotechnology and Genomic Medicine in conjunction with the Georgia Cancer Center. Services are available to all campus investigators as well as off campus investigators. Services offered by the facility include protein identification, labeled and label-free comparative proteomics by Orbitrap mass spectometry, protein/small molecule quantification using targeted mass spectometry (PRM and SRM/MRM mass spectometry), high throughput protein immune-protein assays (384-well Luminex/ELISA assay), etc.
Contact Us
Proteomics Core
Health Sciences Campus
Georgia Cancer Center - M. Bert Storey Research Building
CN4162
706-721-0601
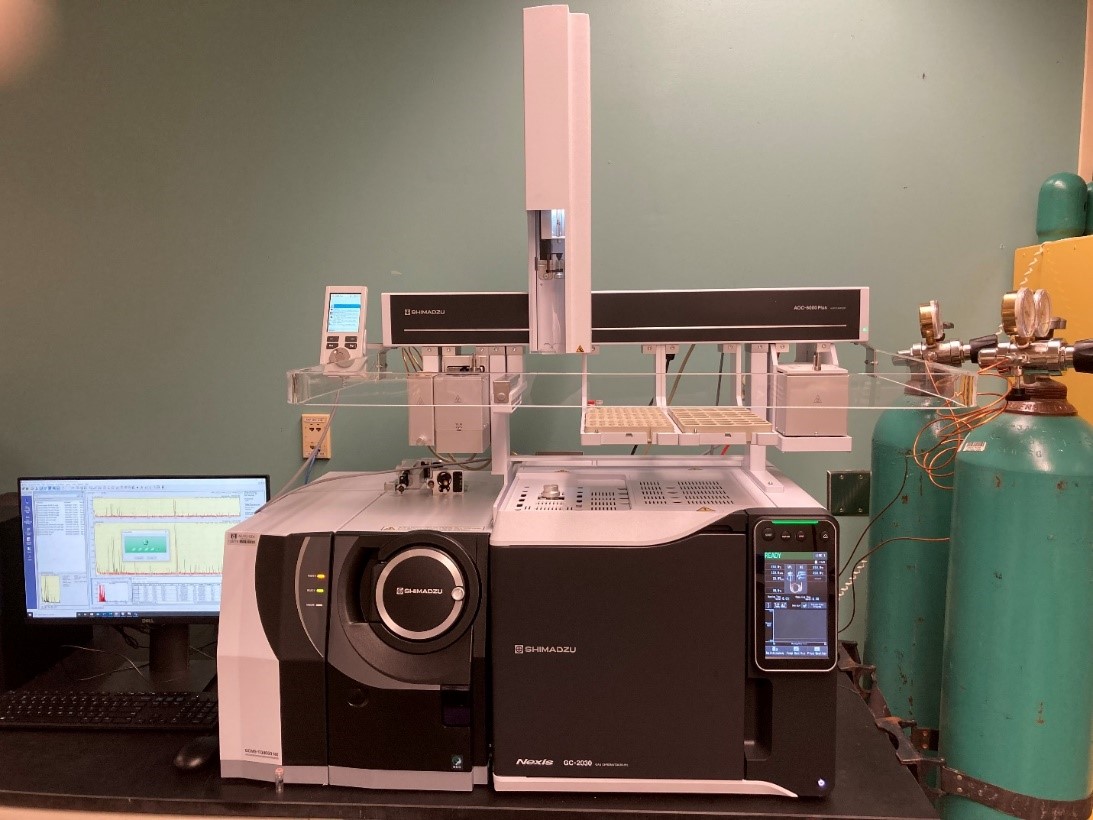
Equipment and Resources
Select image for expanded view
Services
| Area | Analysis Type | Service | Equipment |
|---|---|---|---|
| Proteomics | Untargeted/shotgun analysis | Fast protein identification with LC-MS/LS | Orbitrap Fusion |
| Proteomics | Untargeted/shotgun analysis | Comparative proteomic analysis (Label-free/spctral counting) | Orbitrap Fusion |
| Proteomics | Untargeted/shotgun analysis | Comparative proteomic analysis (TMT/SILAC labeling) | Orbitrap Fusion |
| Proteomics | Untargeted/shotgun analysis | PTM characterization (phosphorylation, acetylation, etc.) | Orbitrap Fusion |
| Proteomics | Untargeted/shotgun analysis | Glycoproteome characterization | Orbitrap Fusion |
| Proteomics | Targeted analysis | Peptide quantification using PRM | TSQ or LC-TQ |
| Proteomics | Targeted analysis | Peptide quantification using PRM | Orbitrap Fusion |
| Metabolomics | Untargeted/shotgun analysis | High resolution metabolome charaterization | Q-TOF or Orbitrap Fusion |
| Metabolomics | Untargeted/shotgun analysis | GC-MS charaterization of metabolome | GC-MS |
| Metabolomics | Targeted analysis | Compounds/metabolites quantification (relative or absolute) | TSQ or LC-TQ |
Meet the Team

Dr. McIndoe received his Ph.D. in immunology and molecular pathology in 1991 from the University of Florida. In 1992, Dr. McIndoe received a Postdoctoral Fellowship to study with Dr. Leroy Hood at the University of Washington in the Department of Molecular Biotechnology. During this five year fellowship, Dr. McIndoe developed his interest in genomics, biotechnology, bioinformatics and automation, culminating in the development of a number of high throughput technologies to automate the genotyping process necessary for complex disease linkage analysis. Following this fellowship, Dr. McIndoe took a senior research scientist position at CuraGen Corporation to develop a high throughput full length cloning facility. In 1999 he returned to academia as an Assistant Professor in the Department of Pathology at the University of Florida where his laboratory focused entirely on bioinformatics and computational biology. In 2002, Dr. McIndoe moved to Augusta University in Augusta, GA as the Associate Director and one of three founding members of the newly created Center for Biotechnology and Genomic Medicine. Over the last 20 years at AU, Dr. McIndoe has been the Director of the Data Coordinating Centers for four national NIH consortia; the Animal Models of Diabetic Complications, the Mouse Metabolic Phenotyping Centers, the Diabetic Complications Consortium and the Innovative Science Accelerator Program. He is currently a tenured full professor in the Medical College of Georgia, College of Graduate Studies and the College of Allied Health Sciences and Director of the Center for Biotechnology and Genomic Medicine.

Dr. Wenbo Zhi received his bachelor degree in Biochemistry from Wuhan University and his Ph.D. degree in Biochemical Engineering from the Chinese Academy of Sciences. He completed his postdoctoral training on proteomic biomarker development for human diseases using LC-MS based technologies at the Center for Biotechnology and Genomic Medicine of Augusta University. After being promoted to assistant professor in 2011, he took the responsibility of managing the Proteomic and Mass Spectrometry Core Facility at Augusta University. In 2022, Dr. Zhi is appointed as the manager of the new consolidated Proteomics and Mass Spectrometry core lab under the leadership of Dr. Richard McIndoe. Dr. Zhi has a broad background and extensive experiences in biochemistry with specific expertise in proteomics, metabolomics, mass spectrometry, high-throughput protein assays, HPLC, etc. Dr. Zhi has been actively involved in the collaboration research on different research topics with both on-campus and off-campus scientists, in addition to his own research on the development of proteomic and metabolomic biomarker development for human diseases, including chronical kidney disease in diabetic patients and acute kidney disease in pre-term infants.

Dr. Xiao earned his Master’s degree in Biochemistry and Molecular Biology from Hubei University, and PhD in Immunology from Wuhan University, China. He has completed his postdoctoral training in the areas of cancer biology and cancer immunology in Georgia Cancer Center. Dr. Xiao has a 3-year research experience in China National Human Liver Proteomics Projects (HLPP) for protein-protein interaction. Dr. Xiao is also interested in the identification of small molecule metabolites, the screening of peptide libraries, and proteomics- & metabolomics-related data analysis.